| UCSC Repeat Browser – A Genome Assembly Hub for Repeat Elements | 您所在的位置:网站首页 › ucscgenome browser › UCSC Repeat Browser – A Genome Assembly Hub for Repeat Elements |
UCSC Repeat Browser – A Genome Assembly Hub for Repeat Elements
|
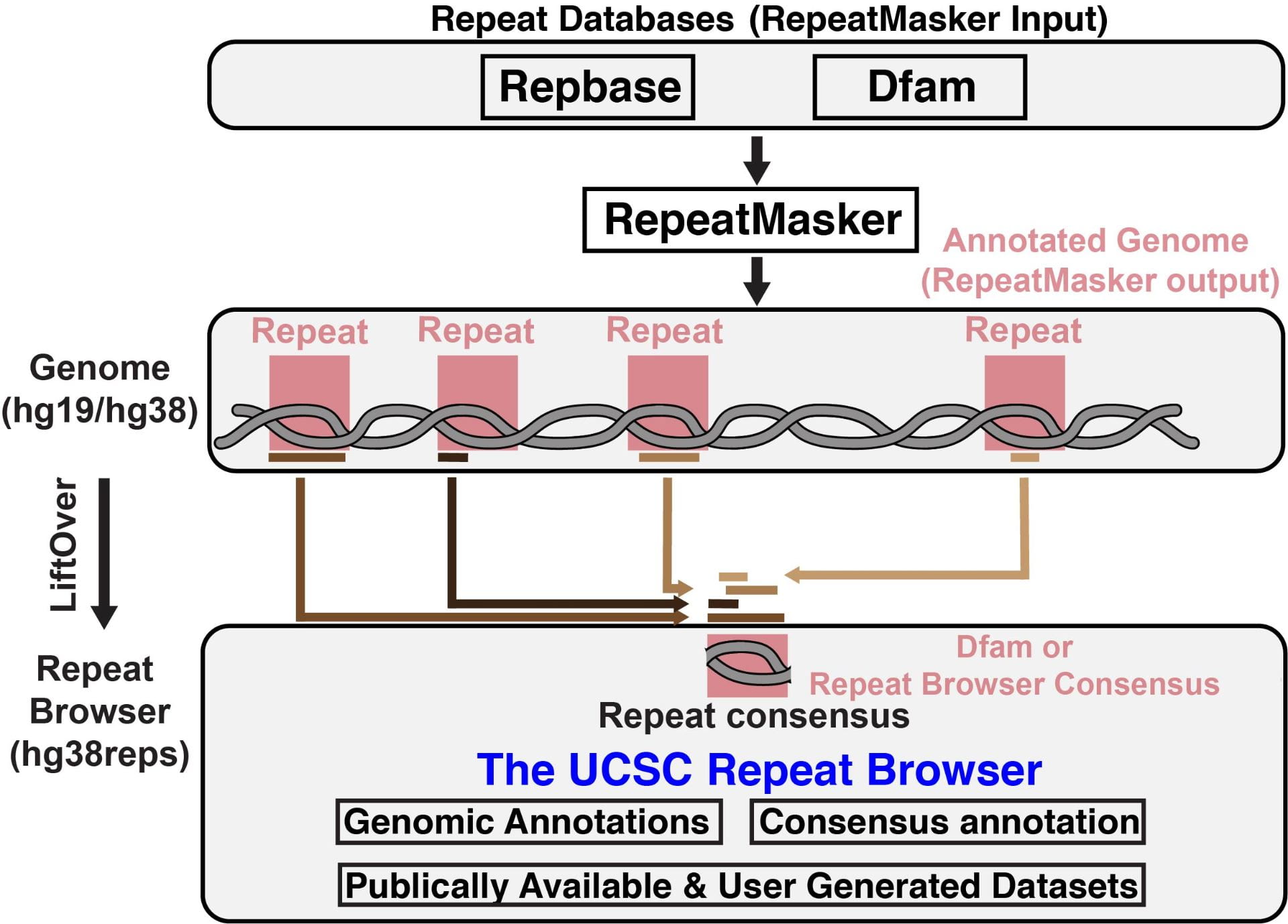
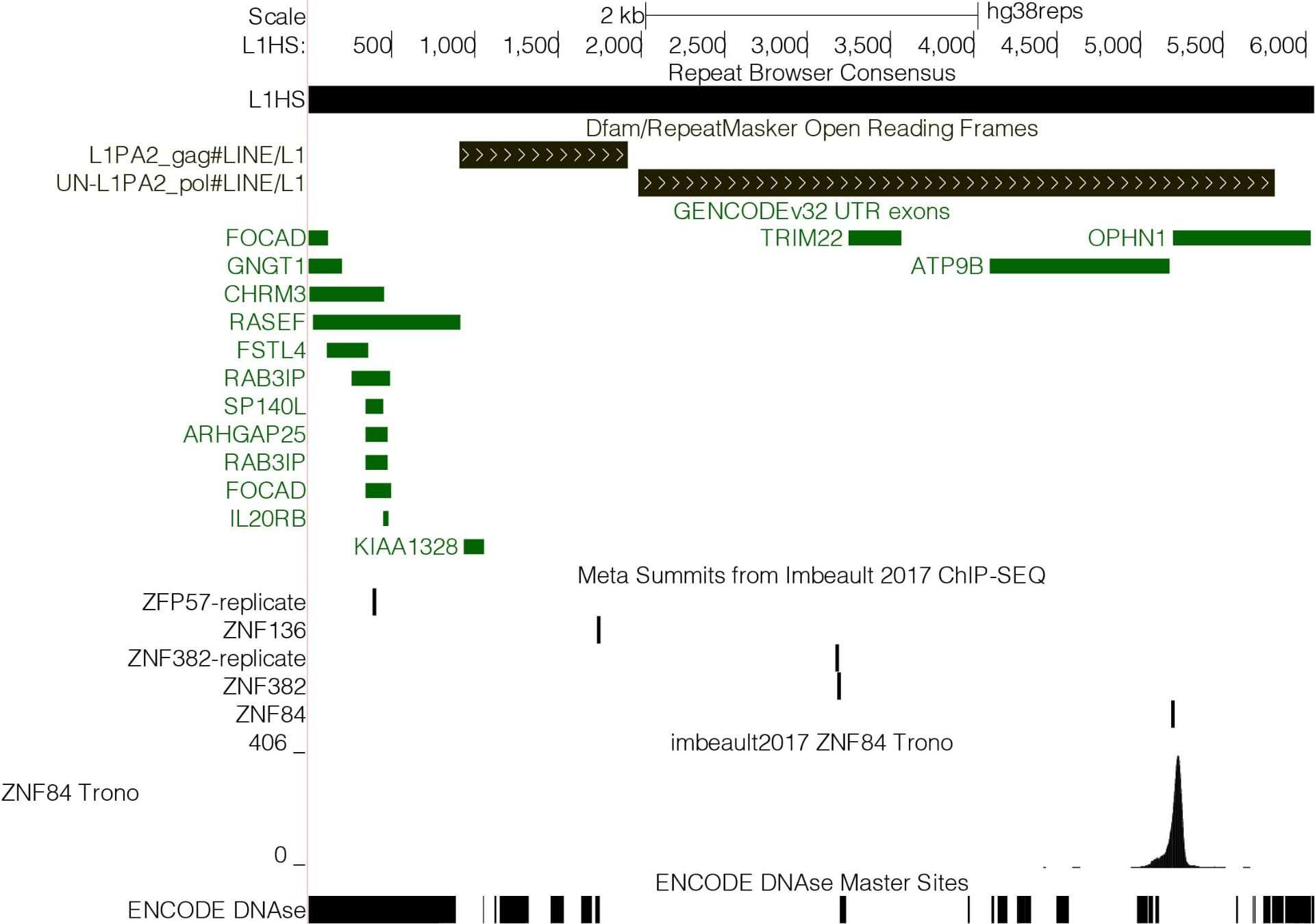
The UCSC Repeat Browser is a genome assembly hub for visualizing genomic data on repeat regions. For our purposes, these repeat regions are equivalent to Transposable Elements (TEs).聽 Data that has been mapped to the human genome (hg19 or hg38) and lies within a region annotated as a TE/repeat (by RepeatMasker) is “lifted” to a consensus version of the TE/repeat.聽 These consensuses come from Dfam or, in some cases, have been built specifically for the Repeat Browser.
The result is an interactive browser environment where “chromosomes” are repeats and data is displayed as tracks, in a manner analogous to a standard genome browser . To learn more, read the paper, explore this website, or start browsing. Please feel free to contact us if you have any questions.Reference: Fernandes, J.D., Zamudio-Hurtado, A., Clawson, H.聽et al.聽The UCSC repeat browser allows discovery and visualization of evolutionary conflict across repeat families.聽Mobile DNA聽11,聽13 (2020). https://doi.org/10.1186/s13100-020-00208-w |
【本文地址】